Electrifying Life Sciences
Electrifying Life Sciences
At the Rosalind Franklin Institute, a specialized team of computer scientists is dedicated to developing software for interacting with large three-dimensional (3D) microscopic biology data sets as part of the Electrifying Life Sciences Project (ELS).
The overall goal of the ELS project at the Franklin is to improve the capability and accessibility of electron cryomicroscopy (cryo-EM). To create 3D pictures of biological samples inside of cells, the cells are cryogenically preserved, imaged using fluorescent light microscopy to find areas of interest, milled to show just those areas of interest and then data is collected. This raw data contains nanometer scale information of cellular compartments and their components (proteins, DNA, RNA, etc). The raw data is next computationally reconstructed into a 3D volume of data and regions within the data are annotated or segmented so that they can be analysed, and the biological question answered. It is very challenging to collect, process, and analyse these large datasets, especially when the analysis workflow includes correlative imaging and segmentation steps as is illustrated in Figure 1.
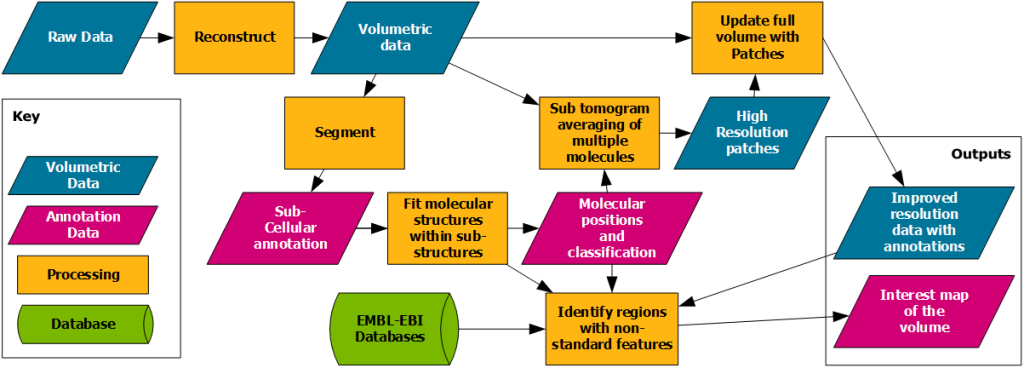
Figure 1. Illustrates the complexity of the data processing pipeline commonly involved in order to generate the relevant outputs.
In the Artificial Intelligence and Informatics (AI&I) group we are automating these workflow steps to solve computationally difficult and time-intensive problems by developing cross platform open-source software tools. We also aim to integrate these with commonly used data analysis and visualisation software such as ImageJ and napari, and EM specialised software such as IMOD. Below, we present some recent examples.
RedLionfish is a package for fast GPU/CPU accelerated Richardson-Lucy deconvolution of 3D optical images. RedLionfish was chosen for the name of this program so that its initials (RL) are the same as Richardson-Lucy who originally developed the algorithm. This tool is useful for removing optical artefacts in 3D microscopy data, leading to clearer and sharper images. This software is available as a plugin for napari (a 3D data visualization application) and it has been included in 3DCT as a data processing tool for correlative microscopy. Speed improvements are powered by using an implementation of FFT written in OpenCL and it is fast enough so that it can be used in real-time alongside of focus ion beam (FIB) lamella preparation as a way to help the researchers find and target specific areas within their specimen.
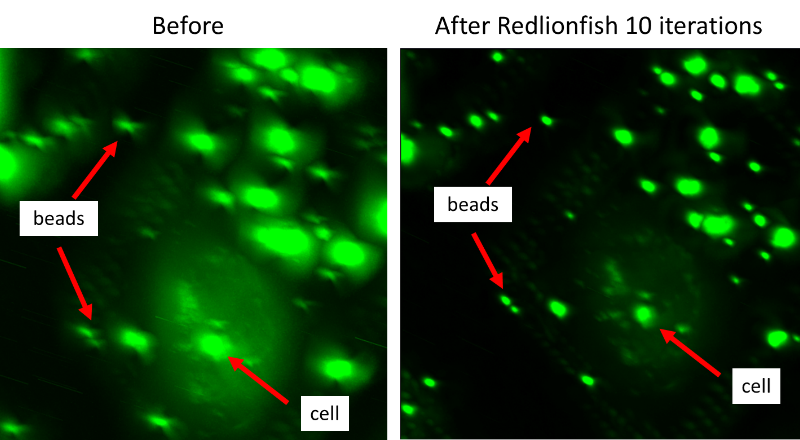
Because we know the expected shape of fluorescent beads (like those shown in figure 2), we can use the 3D point spread function (PSF) to deconvolute the data using the Richardson-Lucy algorithm; and this can be done efficiently due to the accelerated implementation found in the RedLionfish plugin, resulting in an image where beads appear more point-like and biological features appear significantly clearer. Depending on the GPU resources used, this process can take less than a minute, compared to several minutes when using CPU based DeconvolutionLab2 plugin in Fiji/ImageJ giving researchers a more accurate real-time view of their fluorescently marked features during an experiment.
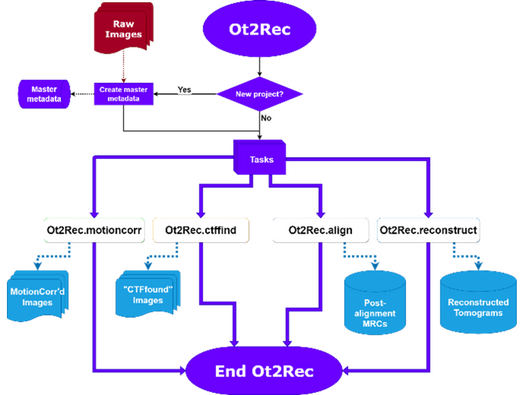
After a feature of interest has been identified and prepared, it is imaged using cryo electron microscopy (cryoEM) where multiple images of the same area are taken at different angles. Next, we have developed a software package called Ot2Rec to automate the reconstruction of these 2D images into a 3D volume. This software is a wrapper for processing packages that are commonly used in the field of cryoEM such as MotionCor2 (motion correction) and CTFFind4 (CTF estimation). Although processing pipeline solutions such as EMAN2 and tomoBEAR already exist, this solution offers advantages by using a general, unified command-line syntax, providing flexibility for future expansion and a more portable codebase. In addition to the above processing packages, Ot2Rec also includes a tool for simulating CTF image stacks and generating 3D point spread functions (PSF) for deconvolution tasks.
Once a dataset has been reconstructed into a 3D volume, the next step is to annotate or segment the data to add meaning to it. SuRVoS2 is a collection of tools to help accelerate annotation and segmentation of large volumetric bio-imaging workflows. It enables either shallow or deep machine learning approaches, using a suite of image processing filters, supervoxels (boundary adherent groupings of similar, adjacent voxels), and annotation hierarchies. SuRVoS2 also provides a set of tools to enable visualization and interaction with large numbers of distributed annotations (e.g. those performed by multiple members of a group or citizen scientists). This application has been implemented both as a napari plugin and as an API for generic programming usage.
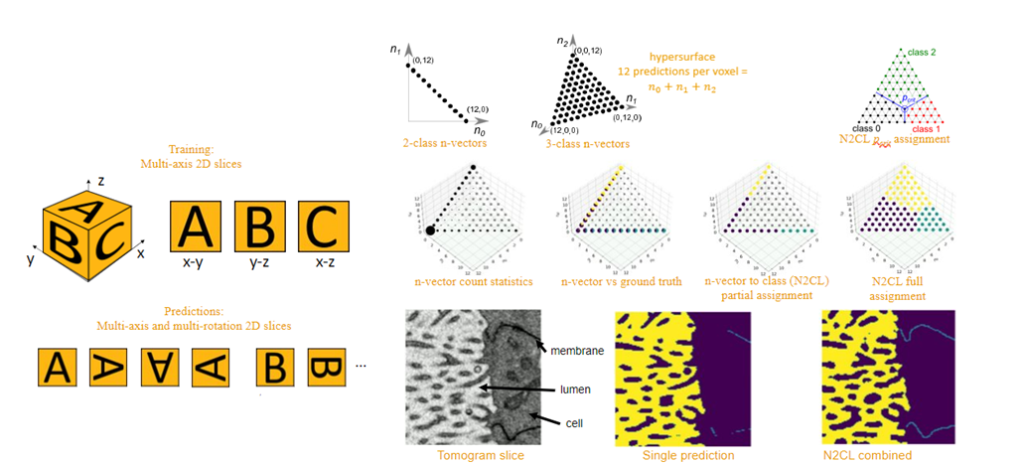
In addition to the options available in SuRVoS2, the AI&I team at the Franklin are working on a more broadly applicable deep learning option for segmentation of very large datasets. Unet+ is a new approach to machine learning training and prediction for segmentation of large volume biological samples that uses the well-known UNet neural network architecture for 2D biological images, but expands to allow predictions of 3D volumes, using a multi-slicing, multi-axis and multi-rotation technique. Computational post-processing methods are currently being developed that combine the multiple generated predictions for optimized confidence metrics. These confidence metrics are important as they give us a way to check that the output from our machine learning algorithms is believable and real.
Together these tools help to speed and automate the collection, processing and analysis of 3D biomedical data, increasing the pace of research at the Franklin and enabling projects which otherwise would not be possible.
Project Leadership
Project Members at the Franklin
Funded by