All-in-One Platform for biomolecule analysis
Researchers from the Rosalind Franklin Institute, Fasmatech and Thermo Fisher Scientific have developed, constructed and characterised a one-of-a-kind bespoke mass spectrometer that is capable of comprehensive characterisation of most types of biomolecules.
The researchers have initially started with peptides and proteins demonstrating near complete sequence information for small proteins (<20kDa) and best in class results for larger proteins; all achieved at competitive speeds and sensitivity allowing access to most types of biological questions.
This research was published last week in the Journal of Analytical Chemistry.
The instrument consists of Thermo Scientific™ Orbitrap™ Exploris™ 480 mass spectrometer coupled to an OmnitrapTM platform which was developed by Fasmatech. This particular design allows several discrete reaction chambers and access to an unmatched range of activation techniques including collision induced dissociation (CID), Infra-red multiphoton dissociation (IRMPD), ultra-violet photodissociation (UVPD) and several electron-based dissociation regimes (ExD). This instrument allows researchers to combine multiple fragmentation techniques within one mass-spectrometry platform either simultaneously or in a consecutive arrangement.
“Another flavour of the omnitrap platform and the most powerful one!” Dimitrios Papanastasiou (Founder, Fasmatech)
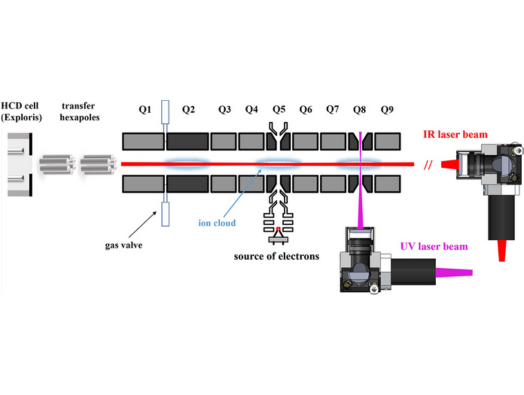
The researchers are primarily interested in pushing the current limits of proteomics workflows. Now that initial characterisation is complete the researchers will be attempting to develop facile approaches for extend protein context information. They will be identifying protein- protein (or -RNA or -glyco or -lipid) interfaces through crosslinking approaches as well as tackle currently challenging proteoforms such as glycoproteins.
“This platform will be of great interest to structural biologists, as it will allow fast and efficient characterisation of proteins and will be complimentary to imaging techniques.” said Dr Nikita Levin, Postdoc at the Franklin and co-first author of the research paper.
Furthermore, to aid automated data annotation, the Omnitrap team will collaborate with Franklin’s AI team to generate fragmentation models using deep learning approaches to, subsequently, build proteoform identification tools.
“The initial data suggests that we have an extremely powerful platform for proteomics, and we are looking forward to putting it to work answering pressing questions in many of the biological systems under study here at the Franklin” said Prof. Shabaz Mohammed, Deputy Science Director at the Franklin, Associate Professor of Proteomics at Oxford University and the paper’s senior author.
Related publication
