Key Biosynthesis Enzyme Structurally Resolved
Franklin researchers, in collaboration with the University of Oxford, Keele University and Queensland University of Technology have succeeded in characterising the bifunctional enzyme NDST1. In doing so, they have closed a knowledge gap: providing the final experimentally resolved structure for the enzyme families involved in the biosynthesis of heparan sulfate.
Heparan sulfates (HS) are an abundant class of polysaccharides (long chain sugars). They are found on cell surface membranes and within the extracellular matrix in the form of HS proteoglycans, where multiple HS polysaccharide chains are bound to a core protein. Because of their abundance and diversity, HS proteoglycans are implicated in a variety of biological processes, such as growth factor signalling, cell adhesion, and host-pathogen binding. Heparin, the widely used anticoagulant medication, is a highly sulfated HS analogue that is also synthesised in mast cells.
Biosynthesis of HS occurs in the Golgi and involves a range of biosynthetic enzymes. The enzyme studied in this paper, NDST1 (N-deacetylase-N-sulfotransferase 1) is the most abundant enzyme within the NDST family and the dominant enzyme in most human tissues. NDST enzymes are particularly important because they are the first to act on the polysaccharide after initial polymerisation. First author, Dr Courtney Mycroft-West explains: “It’s thought that N-sulfation is used as a kind of marker put down for further biosynthetic steps. So other sulfation sites are clustered around where the N-sulfates are initially placed by NDST1 activity.”
There is a great degree of structural variation in the HS found throughout the body. The variable length polysaccharide chains are decorated with different sulfate groups, and the position that these groups are attached during biosynthesis dictates the biological function of the polysaccharide. Ultimately, this determines the ability for different proteins to bind to the resultant proteoglycan during its function.
Using cryo-electron microscopy (cryo-EM), the researchers were able to determine the structure of NDST1 to a nominal resolution of 2.70 Å, enabling modelling of nearly all residues along the protein chain. The model resolved molecular details of both the NDST1 sulfotransferase and deacetylase catalytic domains, shedding light on the mechanisms by which these domains process their polysaccharide substrates. The team were also able to resolve a previously unreported non-catalytic N-terminal domain, which may have potential significance in supporting polysaccharide binding to NDST1.
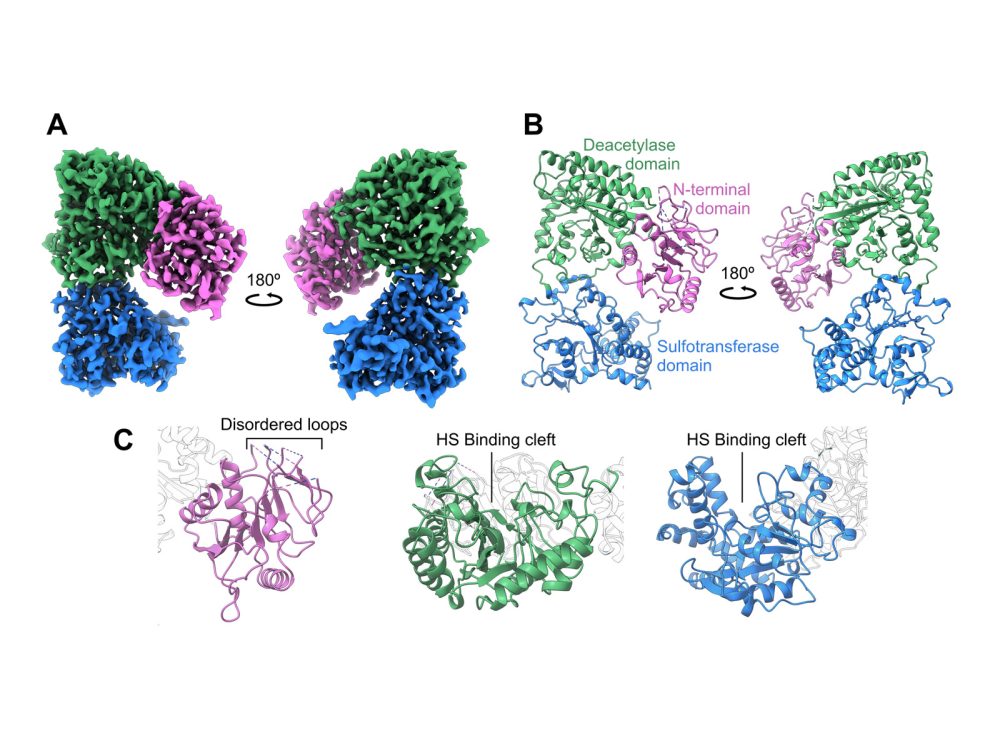
But with the enzyme’s structure came a surprise: the two catalytic domains of NDST1 were facing away from each other, meaning direct domain-to-domain transfer of the HS substrate was unlikely. When the team paired this with the electrostatic surface potential calculated of the enzyme, the sulfotransferase domain surfaced as the likely candidate for initial HS binding, even though deacetylase activity is known to occur first during enzymatic processing. Co-author, Dr Liang Wu comments: “We think that the sulfotransferase domain acts as an anchor to bind to the substrate before subsequent deacetylase activity, thereby providing a mechanism to coordinate the two catalytic domains despite their spatial separation.”
To fully characterise the functionality of NDST1, the researchers also generated a panel of nanobody binders. In doing so, they discovered three nanobodies which modulated the catalytic activity of the enzyme. Of these, two enzyme-nanobody complexes, one inhibitory and one enhancing, were also structurally characterized by cryo-EM. Liang explains: “Understanding how these activity modulating nanobodies bind to NDST1 paves the way to use them in more functional contexts, for example, to control HS biosynthesis in living cells.”
The team are now working to image NDST1 within the Golgi itself, to understand its potential interactions with other enzymes in the HS biosynthesis pathway. “We want to see if we can actually find these complexes.” explains Courtney, “So we’re trying to set up pipelines for cryo-electron tomography, to see whether we can see enzyme interactions in situ.”
But this is easier said than done – with the myriad proteins and enzymes present in the Golgi, finding the right interactions will be difficult. But if complexes do form, they will be larger and easier to spot than NDST1 alone. “At the moment the challenge is trying to identify them. And if they are quite transient, then [our search is complicated by] how often they might appear.”
