‘Native’ radical creation technique opens new doors in protein editing
In the human body, proteins in our cells are often modified after production. A process called glycosylation helps fine-tune our immune systems, while another called ubiquitination helps remove damaged or unwanted proteins from the cell.
These natural processes, however, only scratch the surface of the potential functions and applications of so-called ‘post-translational’ modification (PTM) of proteins.
Researchers at the Rosalind Franklin Institute and the University of Oxford have been exploring chemical methods of generating these PTMs with a view to revolutionising the way organic chemistry interacts with complex human biology.
Their latest technique, outlined in the American Chemical Society’s Central Science journal, demonstrates a way of producing a reactive molecule known as a radical on the protein itself – rather than being introduced separately. These radicals help create carbon-based bonds that pave the way for new functions to be added to, or generated within, the protein.
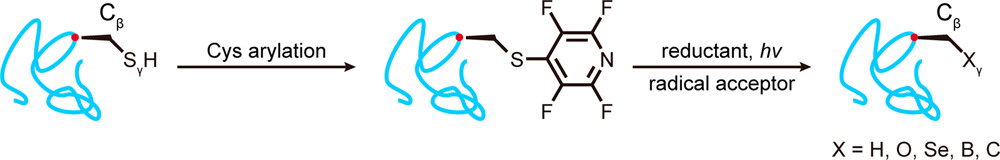
The study builds on the longstanding work of the Franklin’s Next Generation Chemistry theme, which has pioneered light-activated ways of altering the functional information found inside biological molecules such as proteins and sugars.
Medicine is the natural direction for much of this work, with conceivable applications in drug and vaccine development, as well as the therapeutic use of protein editing itself through the enhancement of ‘deficient’ proteins or the introduction of synthetic proteins into the body.
“The word we want to emphasise in this study is ‘native’,” says Dr Yizhi Yuan, a postdoctoral researcher at the Franklin and one of the new paper’s lead authors. “Traditional methods have focused on using an unnatural amino acid called dehydroalanine (DHA) to create the carbon radicals that allow protein modification. We can install plenty of modifications using that approach, but one drawback is that it disturbs what we call the stereogenic centre of the molecule.
“To solve that problem, we generate the radical in situ on the protein backbone, retaining the molecule’s native stereochemistry – its arrangement of atoms.”
Atoms in naturally occurring amino acids – the building blocks of proteins – tend to be structured in what is known as the ‘L’ configuration. The DHA approach to radical generation, however, produces a more unusual ‘D’ configuration that is not recognised by some protein-modifying enzymes, meaning the desired functional modifications can be less efficient and effective. The research team is now working to refine its new technique, which avoids this problem, and make it accessible to scientists more widely.
Dr Xiaping Fu, postdoctoral researcher at the Franklin and another of the paper’s lead authors, says: “Drug delivery is a clear potential application for this technique. For example, you could link a cancer drug to a protein that would recognise and kill tumour cells, as seen in existing antibody-drug conjugates. It could also be used for the labelling of cell surfaces, in which molecules are attached to a cell to help investigate what’s happening in human biological processes.
“But this is just the beginning, and our next stage involves figuring out how to stabilise the on-protein radicals to enhance the efficiency of the method. One of the most important things about this paper is that the methodology provides a toolbox for scientists everywhere to explore the applications of protein modification through chemical techniques.”
