New simplified methods for imaging DNA nanostructures
DNA nanostructures can be changed and improved using different biological and chemical processes, allowing them to be used more accurately in applications such as drug delivery. As a result, imaging functional and multidimensional DNA nanomaterials is important in a range of different contexts, such as nano-mechanics, biomimicry, biocomputing and biomedicine, to name a few.
The images produced of these important nanomaterials have an image contrast which is much lower than those from heavier nanoparticles using conventional transmission electron microscopy (TEM) so 3D scaffolds cannot be resolved in a native state.
Electron ptychography is a unique microscopy technique which is computational and can use iterative algorithms, such as an extended ptychographical iterative engine (ePIE) to create both 2D and 3D images. In Ptychography, a convergent probe is used to illuminate and scan the sample with overlapping illuminating areas. Electron diffraction patterns at different scan positions are recorded and this data is then used to create phase images computationally. The images created can have a resolution that is greater than that in conventional TEM imaging. This is because ptychography can remove the effects of lens aberrations, which limits the resolution of conventional TEM imaging.
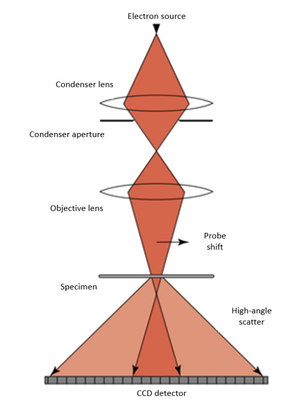
The researchers show that electron ptychographic tomography (EPty-Tomo) can be used to obtain high contrast phase information from DNA origami scaffolds without staining. They collected “4D-STEM” datasets of diffraction patterns from an array of scan positions in relation to the specimen (this is used for many different image processing methodologies). The ePIE algorithm was used to construct the phase data.
In order to reconstruct 3D images, information form the X, Y and Z axis is needed which is not provided by conventional 2D TEM imaging. In tomography, the sample is tilted, and this tilt-series dataset is reconstructed using one of several tomography algorithms (in this case the GENeralized Fourier Iterative Reconstruction (GENFIRE)).
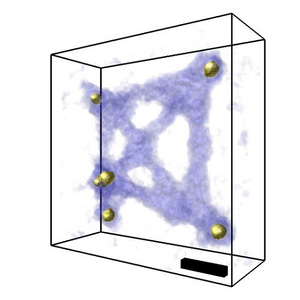
The specimen was tilted at 23 different angles and 4D-STEM datasets were recorded at each, ptychographic reconstruction was then run separately to produce 23 2D phase data. A tomography reconstruction was then carried out on the 23 2D images produced to create 3D images.
Ding et al have shown that it is possible to produce high quality high contrast images of fine structures with a workflow which is significantly simplified in comparison to other methodologies. This methodology also allows them to reduce electron dose and beam damage.
