Scientists find new way of ‘chopping’ proteins chemically
A team of researchers from the Rosalind Franklin Institute and Oxford University has discovered, using cheaply available reagents, a novel and simple way of breaking down proteins by targeting a particular chemical bond. The new method appears to mimic an unusual process that takes place in nature and opens up a host of potential opportunities and applications in biology – including the development of therapeutics for disease that target unwanted or damaged proteins.
In chemistry, as in nature, proteins tend to be broken down in the same way: by cleaving a particular bond – a carbon-nitrogen bond known as the peptidic bond – in the molecule’s amino acid ‘backbone’.
Whether through natural enzymes or chemical methods, this process enables a number of important functions, from human digestion to the detailed study of proteins in the laboratory.
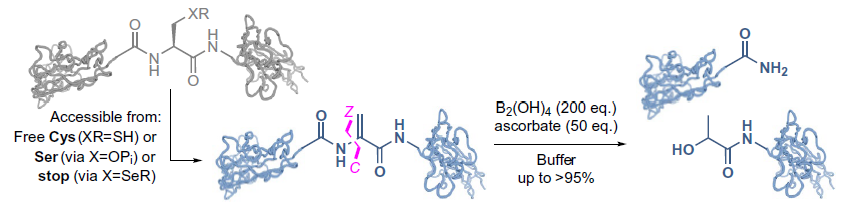
Now, researchers at the Rosalind Franklin Institute and Oxford University have developed a new chemical method of cleaving proteins that uses a boron-based reagent in combination with the addition of an amino acid called dehydroalanine to break the neighbouring nitrogen-carbon bond.
Breaking this nitrogen-carbon bond can also take place in nature, but not often, and mostly for unusual purposes including the production of rare peptides – short chains of amino acids smaller than proteins. These peptides range from those with beneficial effects, such as the peptide-hormone oxytocin, which stimulates contractions during childbirth, to snake venom and other toxins. The research team is keen to explore the open biological question of how nature is able to produce these highly potent peptides in tiny quantities, and whether that process could be replicated in the lab for human benefit.
The new cleavage technique – a first using chemical methods – thus opens up a host of biological opportunities and could even, in theory, be used in disease therapeutics by destroying unwanted proteins or inhibiting their activities.
Professor Ben Davis of the Rosalind Franklin Institute, one of the study’s senior authors, said: ‘When you look at an amino acid residue, there are three places where you can “chop” along the backbone. Pretty much all enzymatic and chemical techniques involve cleaving the carbon-nitrogen bond, which is sometimes referred to as the peptidic bond.
‘What we’ve done is find a way of chopping the next one along: the nitrogen-carbon bond. And instead of producing a carboxylic acid, this method produces an amide molecule.
‘Nature does this in a nifty way to make chemical messengers called neuropeptides, or toxins, or venoms, where for various possible reasons it seems important to use an amide rather than a carboxylic acid. So our chemical technique looks like it’s partially biomimetic of that enzyme-catalysed process.’
To test the potential biological applications of the technique, the research team used it to generate an active neuropeptide within the contents of a cell. They also demonstrated the disruption of the signalling, or communication, system of a common protein called ubiquitin that is implicated in the development of diseases including cancers.
Professor Davis added: ‘This cleavage reaction has an intriguing mechanism and I hope it can provoke lots of thought about what it could be used to do. It’s another example of seemingly esoteric in-cell chemistry, or chemistry applied to proteins, that might in fact drive new and diverse applications in biology.’
The work was led by then-PhD student Dr Tim Mollner, from Oxford University’s Department of Chemistry, in collaboration with group leaders Professor Davis and Professor Shabaz Mohammed (Rosalind Franklin Institute) and Professor Daniel Anthony (Department of Pharmacology, Oxford University).
