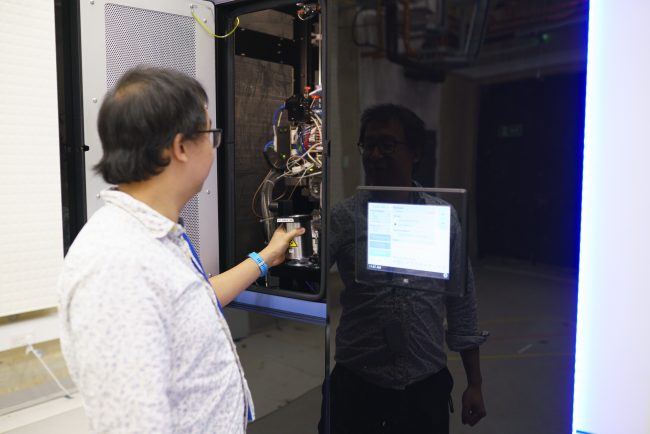
Experimental Automation
We integrate in situ structural biology, computer vision and research software engineering to develop imaging feedback–driven workflows for advanced microscopy. Our goals are to make high-resolution structural methods faster, more robust and accessible.
Cryo-electron microscopy (cryo-EM) and cryo-electron tomography (cryo-ET) allow researchers to visualise the molecular machinery of cells at exceptionally high resolution. In particular, cryo-ET enables imaging directly inside intact cells, preserving the native environment in which molecular complexes operate. This provides a powerful bridge between molecular structure and cellular biology to understand structure in relation to function.
Essential to this process is to thin down cells and tissues to 100-200 nm thin sections termed lamellae, using a (plasma) focused ion beam. Although some automation exists, producing high-quality lamellae is still slow, error-prone, and heavily dependent on specialist expertise.

By integrating computer vision and machine learning into the sample preparation workflow, we monitor lamella quality in real time and adjust milling parameters automatically. This data-driven feedback aims to improve lamella quality, throughput and make the methodology more accessible to non-experts — enabling a wider range of researchers to apply cryo-ET to their biological questions.
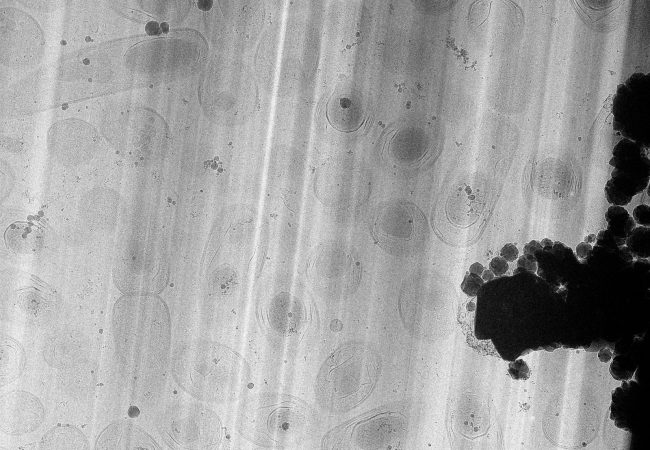
Large Volume Tomography
High resolution large volume tomography with electron microscopy has the potential to transform our understanding of life, by linking the atomic and molecular structure of protein complexes in their biological context – the cell.

Advanced Research Computing
The Franklin develops state of the art digital infrastructure to enable cutting edge research. Our interdisciplinary research generates a variety of data structures and sizes that all need to be stored, curated and processed to give data driven insights.