Capturing Complexity: How AntigenApp Is Transforming Nanobody Discovery at the Franklin
Researchers at the Franklin studying schistosomiasis- a neglected tropical disease affecting more than 200 million people globally- have now observed the structure of a key protein thanks to a unique blend of nanobody discovery and advanced data science.
Cracking the Structure of a Key Schistosome Enzyme
Schistosomiasis is caused by parasitic flatworms called schistosomes, which affects the urinary tract and intestines. Long term infection can cause liver damage, kidney failure, infertility and bladder cancer. Children infected with the parasite may have poor growth and learning difficulties. Schistosomiasis affects more than 200 million people across Africa, Asia and the Americas.
Current treatments for the disease are reliant on mass administration of Praziquental, which does not kill the juvenile parasitic stages and can cause severe side effects.
One enzyme, known as Schistosoma mansoni Cathepsin D1, SmCD1, has been identified as a potential therapeutic target which the Franklin has been studying in collaboration with the London School of Hygiene and Tropical Medicine and the Oswaldo Cruz Institute in Rio de Janeiro.
Kelly Parker, first author on the paper and Research Assistant at the Franklin, said, “SmCD1 is thought to be especially important during the juvenile phase of schistosomes, so could be a good candidate for a more targeted therapeutic. Despite this, previous studies have not been able to fully characterise the structure of the enzyme, which would be a key first step to developing a therapeutic.”
The group of researchers identified a high affinity nanobody that bound to the parasite enzyme and not to closely related human enzymes. By fixing the enzyme in a specific three dimensional shape, the nanobody enabled the team to determine the first crystal structure of SmCD1.
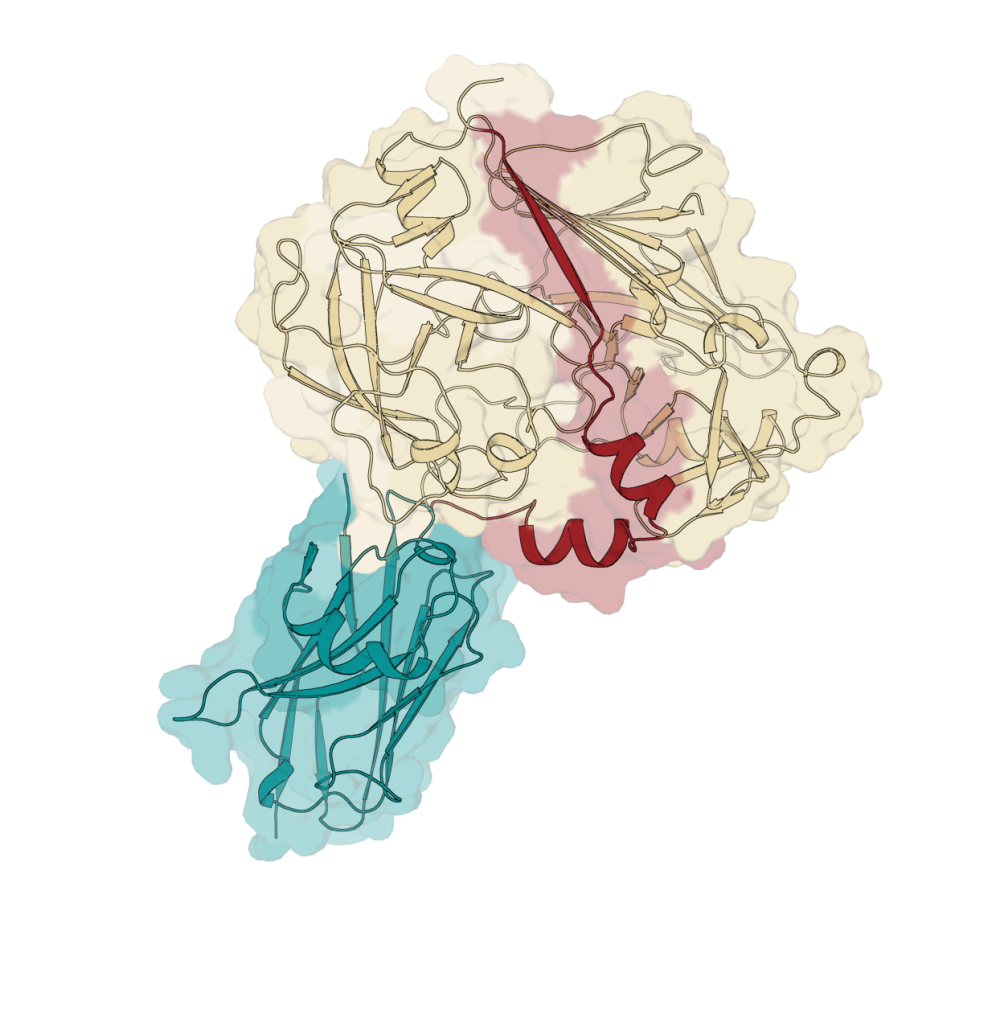
Nanobodies—single‑domain antibody fragments derived from camelid antibodies—are a powerful tool in structural biology, biotechnology, and therapeutics. Their small size, stability, and ease of engineering make them ideal for stabilising difficult proteins such as SmCD1.
Behind the Scenes: AntigenApp Makes Discovery Faster and Smarter
This project utilised new software which supported the entire nanobody generation pipeline. Known as AntigenApp, it was developed by the Franklin Nanobody and Advanced Research Computing teams working closely together and they hope AntigenApp will be useful to the wider scientific community.
AntigenApp captures data from every step of nanobody production, from immunisation to sequence analysis. It integrates powerful bioinformatics tools—including IMGT V‑Quest for automatic sequence annotation and BLASTp for identifying related nanobody sequences—bringing together analysis steps that often require a patchwork of external tools and manual record‑keeping.
“AntigenApp helped us to automate previously time-consuming tasks freeing up time and speeding up we how do things. The development of AntigenApp was a real team effort, working with the ARC team was critical to the success of this project,” said Dr Lauren Eyssen, Staff scientist, Nanobodies Discovery Platform.
Why Data Management Matters More Than Ever
Good scientific practice is rooted in good data management. AntigenApp ensures that experimental metadata, sequences, assay results, and structural insights are all stored consistently and securely in one place. This creates a robust foundation for downstream computational applications.
And the team already has big plans for what comes next. “We’re excited because this data management system will allow us to move on to the next step, where we will incorporate large language models to help us to more quickly identify nanobodies and improve how well they bind to our targets,” said Dr Alex Lubbock, Senior Research Software Engineer, Advanced Research Computing Platform.
A Growing Field with Growing Tools
Lauren Eyssen and Kelly Parker recently discussed the use of nanobodies and innovations in how they are produced in a feature for Nature Methods, read the full feature here. Nanobodies are playing an important role in understanding the molecular basis of diseases caused by viruses, bacteria and parasites. Building this understanding is exactly what the Franklin’s Challenge How Pathogens Interact with Human Cells aims to do. “We are using nanobodies to work out how viruses interact with cells and initiate infection and hope to identify new ways of detecting and treating infectious diseases” said Professor Ray Owens, Science Challenge Lead and Head of the Nanobody Discovery platform at the Franklin.
