Moving from cells into tissue: a new tomography method
Researchers led by Dr Michael Grange at the Rosalind Franklin Institute have developed a robust molecular imaging workflow for brain tissue samples. Their approach utilises plasma ion beams, which increased the size of samples that could be extracted for imaging, and enables researchers to look at samples from multiple individuals or patients. The ability to look at molecular changes occurring at the population level could unlock fundamental new insights.
The work was published this week in Cell Reports Methods.

Molecular interactions are responsible for connectivity, homeostasis and maintenance. Understanding these interactions in a healthy and diseased state could lead us to find new pathways to treatments.
Cryogenic electron tomography (cryo-ET) is a technique that allows researchers to gain detailed structural information in a biologically relevant environment, i.e. inside a cell or tissue, with the potential for atomic level information.
However, it has before now been difficult to prepare tissue samples in a consistent and robust manner for cryoET, with a limit to the size of samples accessible. To achieve this improved workflow, the team have had to improve several aspects of the process from sample preparation to data analysis.
Dr Callie Glynn, co-first author of the paper, said, “I think this paper represents an exciting step forward for the field, it essentially extends our ability to look at 2D cell culture into more 3D systems that might be involved in more complex processes that you can’t necessarily capture with just a monolayer of cells on grids.”
“These more complex systems do mean that we are creating an order of magnitude more data than we would have previously. So once there’s a lot of data, you get to the bottleneck of actually analysing the data and quantifying features there. Working with the AI & I team at the Franklin, we have been able to develop a semi-automated deep learning approach for quantification of features, which has been helpful in speeding up the analysis.”
The team were able to use biopsies from several mice and sample multiple regions of interest within the brain. This included taking samples at different geometries, allowing researchers to adapt their approach depending on the area of interest or cell types they are targeting. Such semi-continuous approaches allow sections of tissue to be assessed in a spatially controlled manner. With their combined approach, specific sub-regions of the hippocampus could be targeted and correlated using micron-scale fluorescence microscopy to nanometre-scale imaging by cryo-ET.
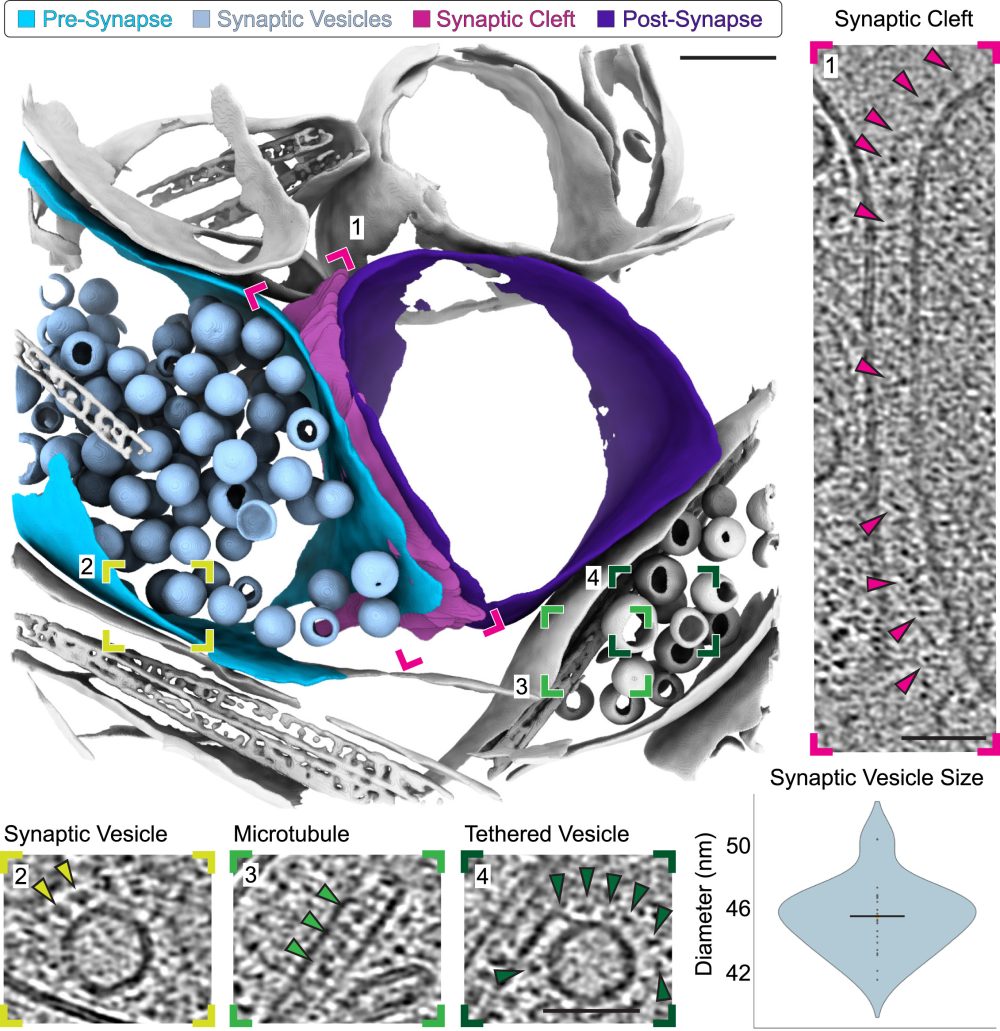
The workflow developed is generalisable to other tissues, so it could equally be used to look at human tissue samples, which is a direction the team hopes to move in the future.
Dr Michael Grange, Tomography group leader at the Franklin and senior author of the paper, said, “What we can do now with the approach we have described in the paper is take 200 μm long tissue chunks straddling several regions of the brain allowing us to trace features throughout, and provide molecular snapshots of these networks. With these snapshots, we can better understand how cellular components are organised, and in the future how this organisation goes wrong in disease.”
